Time-Dependent Genomic Signatures for Cancer Classification and Prediction
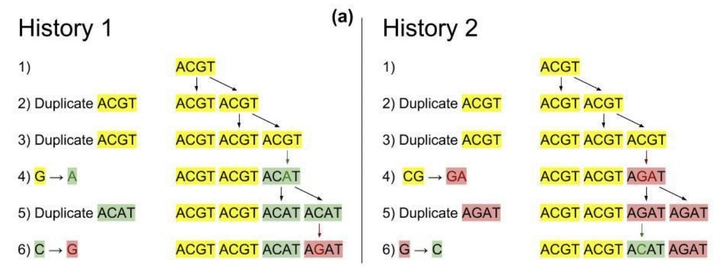
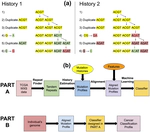
We consider two forms of mutations in the genome:
- Duplication: The formation of adjacent copies of a substring, usually caused by “slipping” during the replication phase.
For example: $T\underline{CG}A \rightarrow T\underline{CGCG}A$ - Point Mutation: The change of a single base pair.
For example: $T\underline{C}GA \rightarrow T\underline{G}GA$.
The genome contains non-coding regions called “repeat regions” with significant duplication activity. When point mutations occur in these regions, those errors are propagated through further duplications. Hence, it is possible to partially construct a duplication and point mutation history of the genome using a single snapshot of these repeat regions. Such a history gives insight into the relative rates of duplication and point mutation for specific locations on the genome. We call this history a mutation profile for which we have a patent. We have demonstrated that cancer patients have unique mutation profiles in their non-tumorous DNA, seemingly predicting their propensity for specific types of cancer.