Abstract
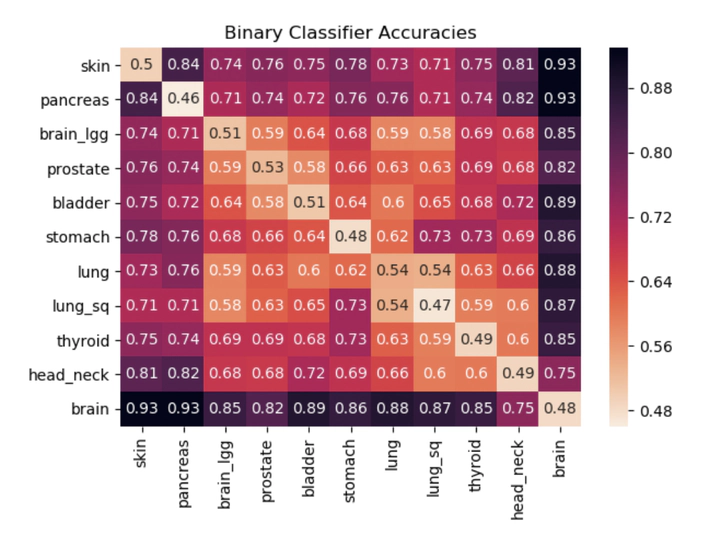
The genome is traditionally viewed as a time-independent source of information; a paradigm that drives researchers to seek correlations between the presence of certain genes and a patient’s risk of disease. This analysis neglects genomic temporal changes, which we believe to be a crucial signal for predicting an individual’s susceptibility to cancer. We hypothesize that each individual’s genome passes through an evolution channel (The term channel is motivated by the notion of communication channel introduced by Shannon1 in 1948 and started the area of Information Theory), that is controlled by hereditary, environmental and stochastic factors. This channel differs among individuals, giving rise to varying predispositions to developing cancer. We introduce the concept of mutation profiles that are computed without any comparative analysis, but by analyzing the short tandem repeat regions in a single healthy genome and capturing information about the individual’s evolution channel. Using machine learning on data from more than 5,000 TCGA cancer patients, we demonstrate that these mutation profiles can accurately distinguish between patients with various types of cancer. For example, the pairwise validation accuracy of the classifier between PAAD (pancreas) patients and GBM (brain) patients is 93%. Our results show that healthy unaffected cells still contain a cancer-specific signal, which opens the possibility of cancer prediction from a healthy genome.