Research
The age of big data promises to revolutionize science and engineering, but it suffers from two critical complications. First, big data introduces heterogeneity from diverse populations that can obscure causality within spurious correlations. Second, rich data gives only a fine-grained picture of the latent abstractions we use to understand the world. I study these issues through the framework of causal inference, which seeks to replace controlled experiments with mathematics on observational data. I’m especially interested in using causal mathematics to answer questions that cannot be addressed by experimentation alone.
Topics
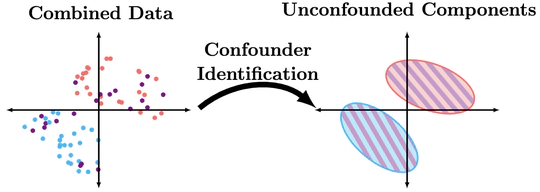
All unsupervised techniques rely on distributional assumptions in order to recover components or clusters. Discrete data poses an interesting challenge in that categorical distributions are inherently non-parametric, prohibiting the use of parametric assumptions. One alternative approach is the use of causal structures to help separate unconfounded components in data. This perspective expands the notion of causal identifiability, as many graphically unidentifiable relationships can be identified.
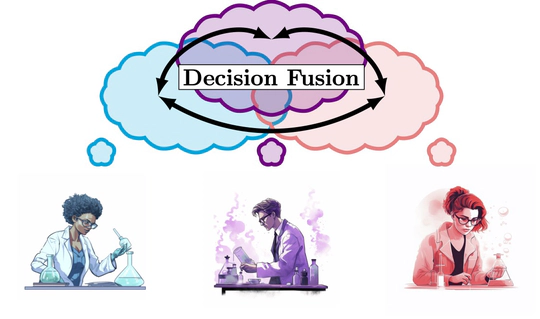
It is not unusual for studies and experts to disagree with each other. Such disagreement is often driven by differing contexts rather than incorrect deductions or bad data. One way to investigate contradiction is to use merged data to build a larger picture. Unfortunately, many private medical settings deny direct access to data. Through decision fusion, I seek to understand when results from different settings are in conflict and what these disagreements can indicate about the underlying system.
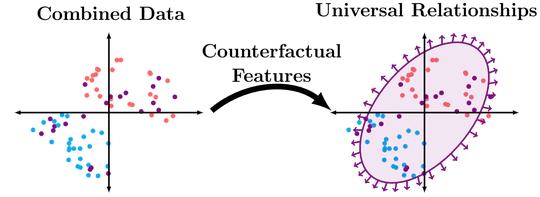
Features often contain a mixture of “good” and “bad” information. From a fairness standpoint, SAT scores contain information about both inherent academic ability, and also access to tutoring resources. From a domain adaptation standpoint, some information may have stable and reliable relationships with the prediction label, while other relationships break down. My work uses insights from causal inference to determine data-representations that sort between the different components of information that are hidden in these ambiguous features.
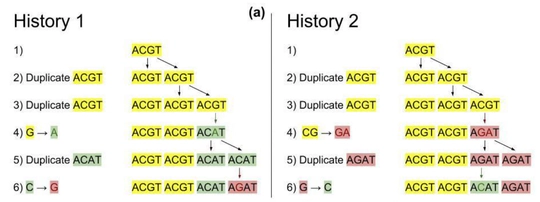
We process non-coding regions of the genome which contain duplication and mutation signatures. These mutation profiles have been shown to be predictive of various forms of cancer.